Image registration and fixel-based analysis now available in MRtrix!
We are proud to announce that MRtrix now includes commands for image registration and fixel-based analysis!
Image registration
The new mrregister command can be used to perform robust rigid, linear and non-linear registration. In a similar vein to Avants et al. (2008) images are symmetrically aligned to a ‘midway’ space. See mrregister --help for complete details.
While mrregister can align any two 3D or 4D images, it has been specifically designed for registration of Fibre Orientation Distributions (FOD). If the input images contain a spherical harmonic series, then FOD registration will be automatically performed and include the required FOD reorientation.
The current version of mrregister is limited to using a mean squared intensity metric, and therefore it requires input images to be in the same intensity range. However we intend to include a normalised cross-correlation metric for both linear and non-linear registration in the near future.
mrtransform
We have also made significant changes to the mrtransform command, so that it can now apply both the forward and reverse linear and non-linear transformations generated from mrregister (including transformation to the midway space). Like mrregister, the mrtransform command will automatically detect if the input is a spherical harmonic series and perform FOD reorientation. In addition, modulation of FODs can be optionally applied to preserve the total fibre density across a fibre bundle’s width.
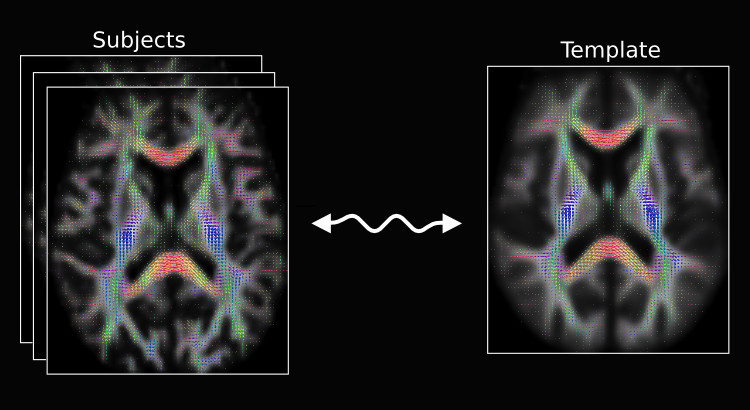
Population template script
The recent update also includes a python script called population_template for building an unbiased study-specific template using an iterative averaging approach. The script can be used on either scalar 3D or FOD images. Using the -rigid option will ensure the initial linear alignment is rigid, which is suited for intra-subject registration of longitudinal data. While not a requirement, we recommend users use the -mask_dir option to supply brain masks for the input subject images, as this will reduce computation time substantially.
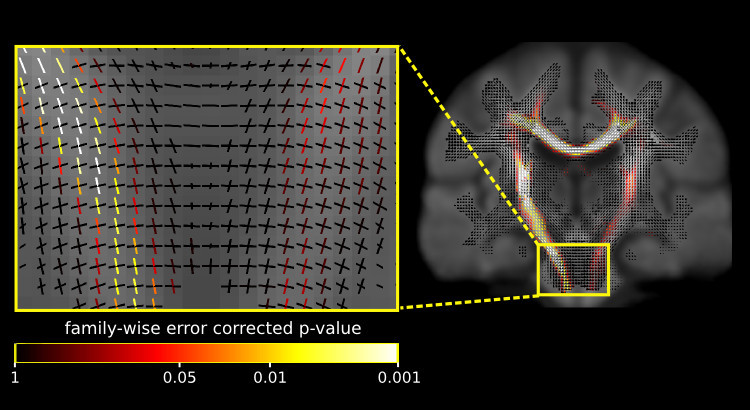
Fixel-based analysis.
This update also includes a number of commands to perform a fixel-based analysis. What’s a fixel? It’s just a fancy word for a specific fibre population in a voxel (a.k.a a fibre bundle element) - see here for a more detailed description. Group strudies using traditional voxel-based analysis permit white matter changes to be localised to a voxel. However, in a fixel-based analysis individual fixels are compared across individuals, which enables significant differences to be localised to a specific fibre pathway, even in regions containing crossing fibres.
For more information on fixel-based analysis and group statistics on fixel images see our paper on connectivity-based fixel enhancement and our upcoming paper on fixel-based morphometry (under review).
Step by step instructions for DWI preprocessing and fixel-based analysis can be found in the MRtrix documentation.
List of new commands
-
mrregister: performs rigid, linear and non-linear registration of images -
fixelreorient: Reorient fixel directions based on the Jacobian (local affine transform) in a non-linear warp -
fixelcorrespondence: After spatial normalisation, assign/match fixels from one image (typically a subject) to another image (typically the template). -
fixellog: A simple command to take the natural log of fixel values. Will likely be incorporated intofixelcalcin the future -
mraverageheader: Estimates the orientation of the average image grid. Used to obtain an unbiased image grid for alignment of subjects to a template. -
mrcheckerboardmask: Used to inspect image alignment after registration. -
mrmetric: Compute the similarity between two images -
warp2metric: Output a map of Jacobian matrices, Jacobian determinats or fibre cross-section for the analysis of white matter morphology. -
warpconvert: Convert between different warp formats -
scripts/population_template: Generate an unbiased population template -
scripts/dwiintensitynorm: Perform a global intensity normalisation of multiple DWI images using the median white matter b=0 value.
Cheers, Dave